Research
The mutations bacteria acquire as they divide provide a transcript of their behavior.
We analyze these mutations in microbial pathogens and commensals to reconstruct past transmission events and identify genes that evolve under natural selection, which represent the most critical challenges for survival and spread of the microbe. We aim to understand those key mutational events that enable infectious diseases on dramatical different time scales from a few weeks to thousands of years.
Located in the heart of Berlin and part of the Max Planck Society we have excellent opportunities for collaboration and access to facilities to nurture our science in every way possible. At the Key lab we are committed to perform science with experimental rigor and curiosity, while creating together an environment that enables each of us to thrive and achieve our individual goals.
Humans are colonized by trillions of microbes, the microbiome, which generates billions of mutations every day - an enormous adaptive potential.
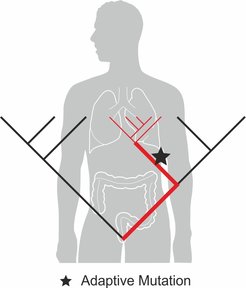
While poorly understood, bacteria might exploit this adaptive potential to translocate into new niches of the human microbiome where they contribute to disease. We have discovered that bacteria colonizing skin lesions of atopic dermatitis patients show a pattern of spatiotemporal spread in agreement with adaptive evolution and we identified adaptive mutations that we confirmed in vitro Key et al. Cell Host & Microbe. The Key lab develops and extends evolutionary inference methods that allow for a detailed reconstruction of the within person transmission and adaptation during disease initiation and progression. We use high throughput single-colony sequencing of target species from clinical samples during the course of disease. The high definition data allows us to develop and test new hypotheses, which holds the promise for successful translation into improved therapy.
Uncovering the deep evolutionary history of infectious microbes is critical for our understanding about past disease emergence and future prevention.
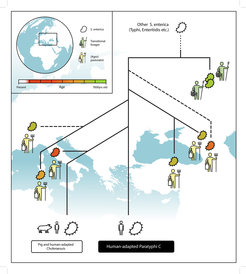
Ancient bacterial DNA recovered from archaeological specimen allows us to travel back in time and directly trace the evolution of important pathogens Key et al. Trends in Genetics (2017). Many human pathogens today are believed to have emerged during the agricultural transition within the last 10,000 years, where major changes in human lifestyle facilitated zoonotic host jumps to humans and subsequent disease. Recently, we provided the first ancient DNA evidence in support of that hypothesis by showing that precursors of a human-specific pathogen emerged in the context of early agriculturalists, which caused infections across Western Eurasia during the last 5,500 years Key et al. Nature Ecol & Evol (2020). A major challenge in ancient pathogenomics is to unscramble the ancient microbial DNA from modern contamination. We have developed first computational tools dedicated to the identification of truly ancient pathogens Huebler*, Key* et al Genome Biology (2019). The Key lab builds upon these results and aims to develop and improve methods for the analysis of ancient microbial DNA and its integration with modern microbial information. We apply these techniques to existent as well as newly generated data to elucidate the timing and genetic changes associated with disease emergence and spread in order to disentangle the deep evolutionary history of microbial pathogens intertwined with the human past.
Selected Publications
- Key et al. On-person adaptive evolution of Staphylococcus aureus during treatment for atopic dermatitis. Cell Host & Microbe (2023) Read News & Views
- Key et al. Emergence of human-adapted Salmonella enterica is linked to the Neolithization process. Nature ecology & evolution (2020) Read Free2read Science News & Views TWiM podcast
- Huebler*, Key* et al. HOPS: Automated detection and authentication of pathogen DNA in archaeological remains. Genome Biology (2019) Read
- Key et al. Human local adaptation of the TRPM8 cold receptor along a latitudinal cline. PLoS Genetics (2018) Read NewScientist TheTimes "Highly Recognized" PLoS Gen Research Prize 2019
- Key et al. Mining metagenomic data sets for ancient DNA: recommended protocols for authentication. Trends in Genetics (2017) Read
- Key et al. Human adaptation and population differentiation in the light of ancient genomes. Nature Communications (2016) Read
- Key et al. Selection on a variant associated with improved viral clearance drives local, adaptive pseudogenization of interferon lambda 4 (IFNL4). PLoS Genetics (2014) Read
- Key et al. Advantageous diversity maintained by balancing selection in humans. Current Opinion in Genetics & Development (2014) Read


